Onco-Seg
Adapting SAM3 for Medical Image Segmentation
Multi-modal AI for tumor & organ delineation across CT, MRI, Ultrasound, and more
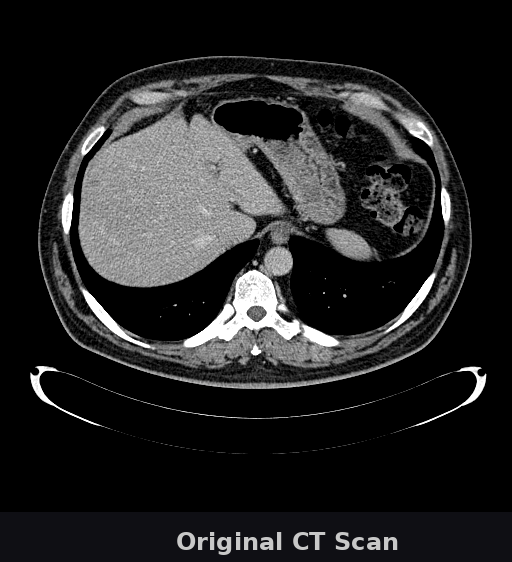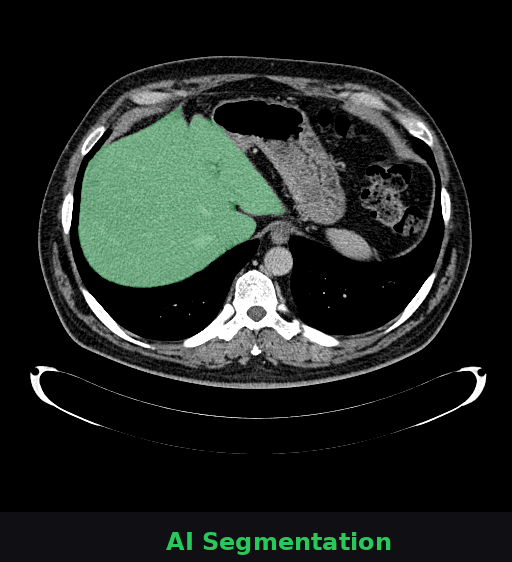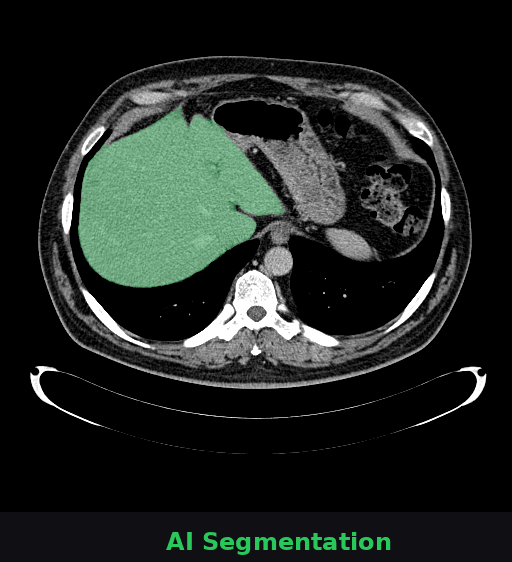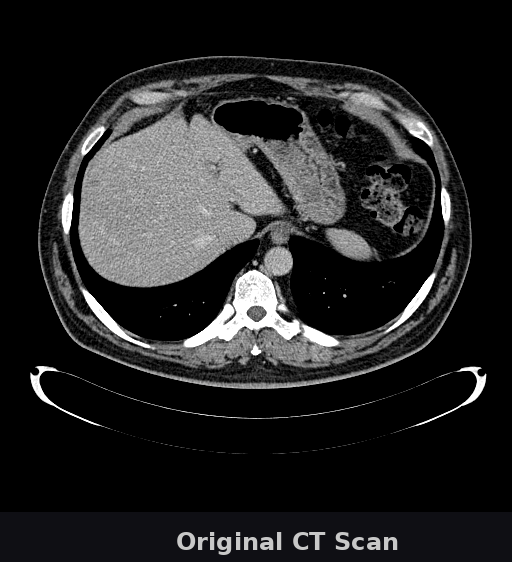
🔬 What you're seeing
This animation shows a CT scan of a patient's abdomen. The AI automatically identifies and highlights the liver (shown in green). This task—which typically takes radiologists several minutes per scan—is completed by Onco-Seg in under a second. Accurate organ segmentation is critical for radiation therapy planning, surgical navigation, and treatment monitoring.
Brain Tumor Segmentation Demo

🧠 Brain Tumor Segmentation
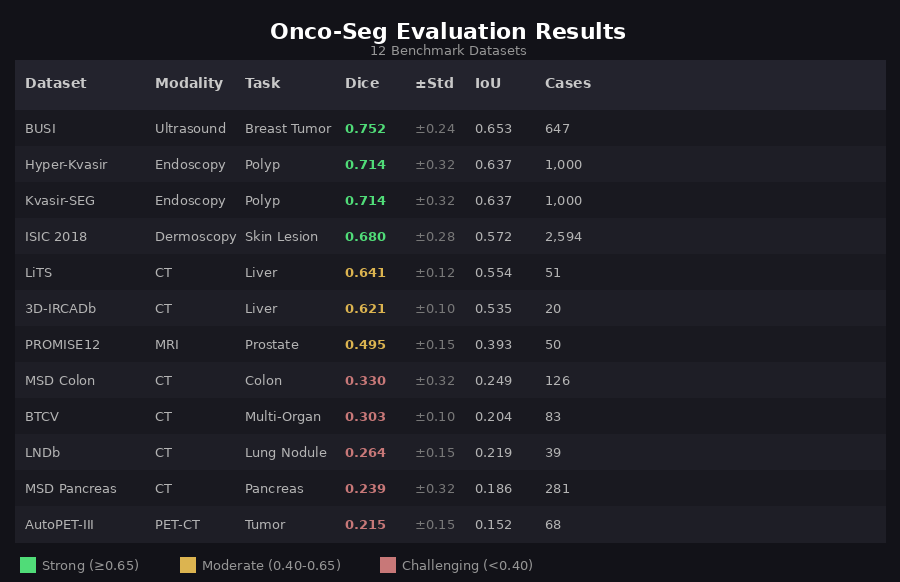
This demo shows Onco-Seg segmenting a glioblastoma from a BraTS MRI scan using the interactive viewer. The AI user types "tumor" as a text prompt, clicks "Find It" to segment the current slice, then "Segment Entire Scan" to propagate the mask across all slices — significantly speeding up segmentation & contouring workflows, largely done manually today. The custom interactive DICOM viewer in the video shows yellow for tumor core and Manual tumor and organ contouring remains a major bottleneck in cancer care. Radiation oncologists spend 2-4 hours per patient drawing boundaries around 30+ organs-at-risk. Radiologists manually measure tumors for treatment response. This process is time-consuming, variable between experts, and difficult to scale. Onco-Seg adapts Meta's SAM3 (Segment Anything Model 3)—trained on billions of natural images—for medical imaging. Using parameter-efficient fine-tuning (LoRA), we transfer SAM3's visual understanding to medical scans while training on just a fraction of the parameters. Onco-Seg was evaluated on 12 public benchmark datasets spanning 6 anatomical regions. Results show strong performance on breast ultrasound, polyp detection, and liver CT, with room for improvement on challenging targets like lung nodules and pancreatic tumors. A single unified model handles CT, MRI, ultrasound, dermoscopy, and endoscopy—no need for separate specialized networks for each modality. Sub-second segmentation on a single GPU enables real-time clinical use, from interactive radiology assistants to automated contouring pipelines. LoRA fine-tuning updates <5% of SAM3's 848M parameters, preserving pretrained knowledge while adapting to medical imaging. Two deployment patterns: interactive "sidecar" for diagnostic radiology (OHIF/PACS integration) and "silent assistant" for automated radiation oncology contouring. Interactive Sidecar: Radiologist clicks on a lesion → instant segmentation with auto-computed volume and diameter for structured reporting. Silent Assistant: CT scan triggers automatic segmentation of 30+ organs-at-risk → DICOM-RT Structure Set → ready for treatment planning. 80-90% time savings. Architecture: SAM3 (848M parameters) with LoRA adapters (rank=16, alpha=32) on attention layers Training: Sequential checkpoint chaining across 8 phases, starting from Medical Segmentation Decathlon (MSD) and expanding to BraTS brain tumors, breast imaging, chest X-rays, and specialized oncology datasets Loss: Combined Dice + Focal loss with modality-specific weighting for class imbalance Infrastructure: 4× NVIDIA RTX 4090 GPUs, PyTorch Lightning, Weights & Biases tracking Onco-Seg is available as a napari plugin for interactive medical image segmentation. The plugin provides a graphical interface for clinicians and researchers who prefer point-and-click workflows over command-line tools. Click-to-segment with point prompts or draw bounding boxes. See results instantly overlaid on your medical image. Point prompts (click on target) and box prompts (draw rectangle)—text-based prompting planned for future releases. Save segmentation masks as NIfTI (.nii.gz) for research or DICOM-RT Structure Sets for radiation oncology treatment planning systems. Segment one slice, propagate to the entire volume. The plugin handles slice-by-slice inference with centroid tracking. The napari plugin is included in the main repository: The plugin supports automatic checkpoint download from HuggingFace, with pre-trained models for general-purpose segmentation as well as specialized checkpoints for breast, liver, and brain imaging. Want to create your own demo video? See our detailed step-by-step guide for recording brain tumor segmentation demos using the napari plugin. This work was supported by the Koita Centre for Digital Health at Ashoka University (KCDH-A). We thank RunPod for GPU infrastructure and Weights & Biases for experiment tracking. Deep thanks & gratitude to: 1. Meta AI & the SAM Team: Special thanks to Meta and the entire SAM team, led by Nikhila Ravi (Meta AI), for being torchbearers of research and innovation in this field with their prolific releases of SAM, SAM2, and SAM3. More importantly, we thank them for making a conscious choice to embrace open source and releasing detailed technical reports and open weights for all SAM releases. We believe innovation in ML & AI at large, and in biomedical & cancer informatics specifically, can truly be accelerated by standing on the shoulders of giants. 2. Bo Wang Lab: The brilliant Bo Wang (@BoWang87) and his prolific lab have been an inspiration. Their pioneering MedSAM work demonstrated the potential of adapting foundation models for medical imaging and paved the way for projects like Onco-Seg. 3. NCI, CBIIT & TCIA: We are deeply grateful to the National Cancer Institute (NCI), its Center for Biomedical Informatics and Information Technology (CBIIT), and The Cancer Imaging Archive (TCIA) for creating such a wonderful open-access resource that has enabled countless research innovations in medical imaging AI. Special thanks to Justin Kirby at TCIA for helping debug minor data access issues and for consistently encouraging innovation built on top of TCIA's datasets. The availability of open-source datasets greatly accelerated our progress on this project. We hope that as research in biomedical machine learning and AI progresses, there is an even greater emphasis on building and releasing open datasets—as the success of AlphaFold has so aptly demonstrated—for the greater public good. We also thank the creators of benchmark datasets (Medical Segmentation Decathlon, BraTS, LiTS, ISIC, Kvasir-SEG, PROMISE12, BUSI, and others). Contact: Ashish Makani — ashish.makani@ashoka.edu.inWhy Onco-Seg?
Supported Modalities
Evaluation Results
Key Features
One Model, Many Tasks
Fast Inference
Parameter Efficient
Clinical Deployment Ready
Clinical Use Cases
Diagnostic Radiology
Radiation Oncology
Technical Details
napari Plugin
Interactive Segmentation
Multiple Prompt Types
Multi-Format Export
3D Propagation
Installation
Recording a Demo
Get Started
Citation
```bibtex @article{makani2026oncoseg, title={Onco-Seg: Adapting Promptable Concept Segmentation for Multi-Modal Medical Imaging}, author={Makani, Ashish and Agrawal, Anjali and Agrawal, Anurag}, journal={arXiv preprint}, year={2026} } ```Acknowledgments
Last updated: January 2026